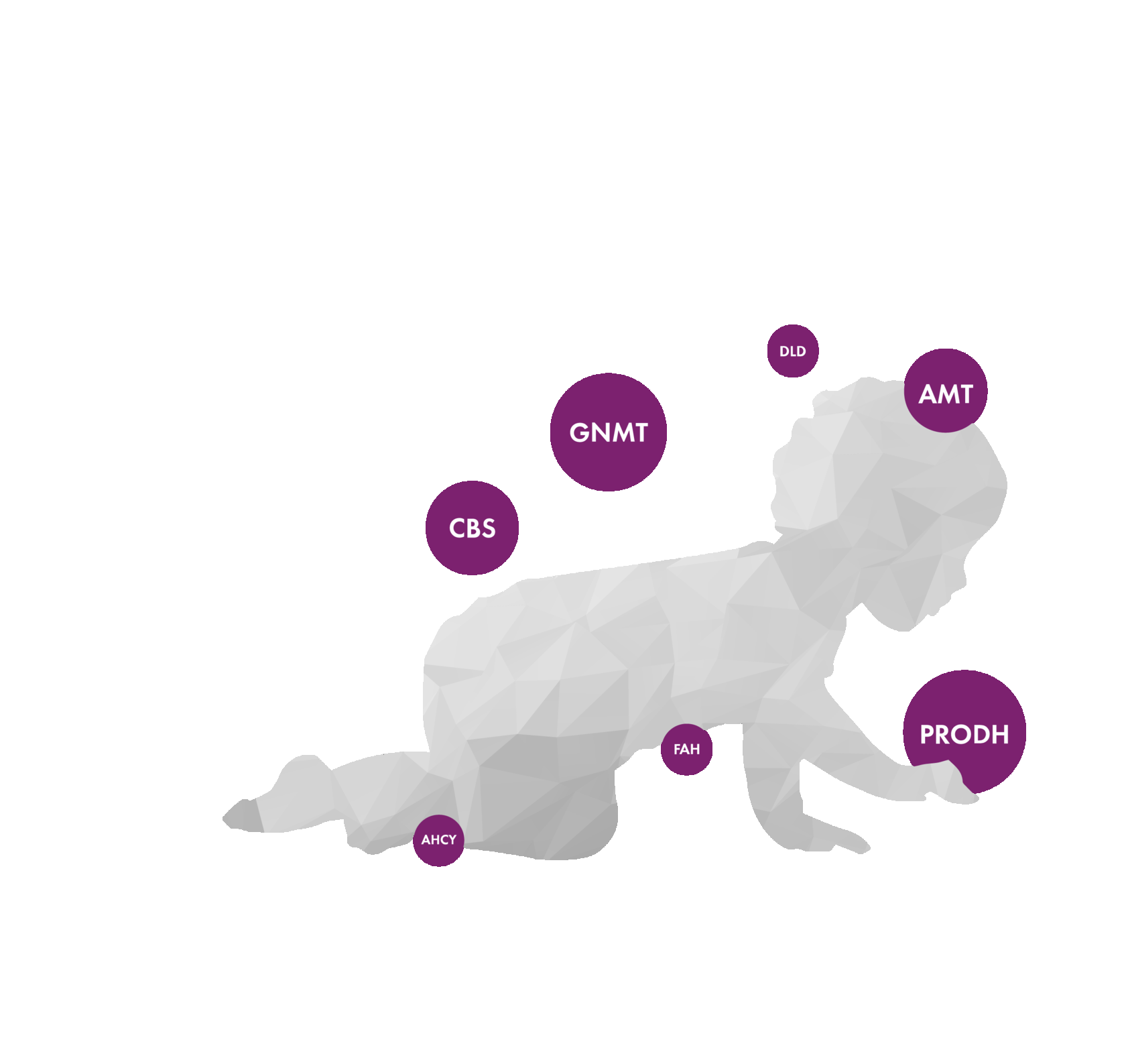
Hypermethioninemia with deficiency of S-adenosylhomocysteine hydrolase
Argininosuccinic aciduria
Maple syrup urine disease, type Ia
Maple syrup urine disease, type Ib
Homocystinuria, B6-responsive and nonresponsive types
Maple syrup urine disease, type II
Dihydrolipoamide dehydrogenase deficiency
Hyperphenylalaninemia, mild, non-BH4-deficient
Dystonia, DOPA-responsive, with or without hyperphenylalaninemia, Hyperphenylalaninemia, BH4-deficient, B
Glycine N-methyltransferase deficiency
Maleylacetoacetate isomerase deficiency
Methionine adenosyltransferase deficiency, autosomal recessive
Homocystinuria due to MTHFR deficiency
Gyrate atrophy of choroid and retina with or without ornithinemia
Ornithine transcarbamylase deficiency
Pyruvate carboxylase deficiency
Hyperphenylalaninemia, BH4-deficient, D
Maple syrup urine disease, mild variant
Hyperphenylalaninemia, BH4-deficient, A
Hyperphenylalaninemia, BH4-deficient, C
Hyperornithinemia-hyperammonemia-homocitrullinemia syndrome
Lysinuric protein intolerance
Dystonia, dopa-responsive, due to sepiapterin reductase deficiency